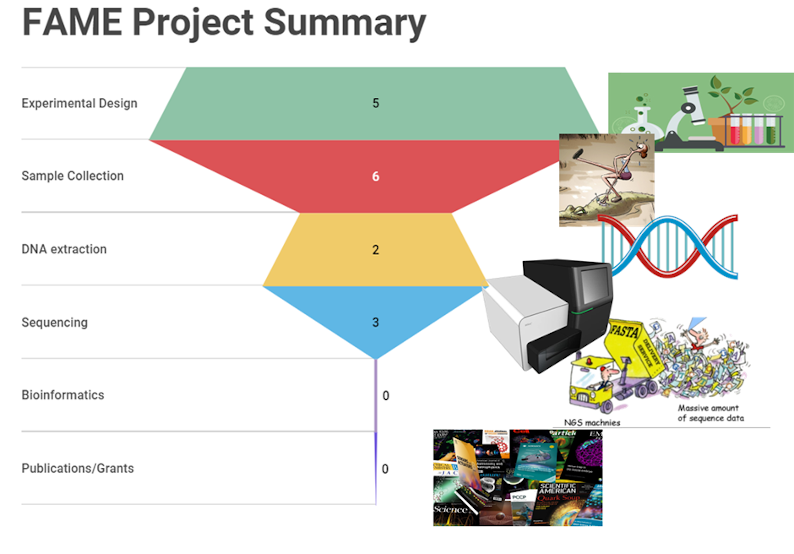
FAME seed projects for 2021
- Environmental DNA and white shark occurrences
- Ecophysiology of pitcher plant microbiomes
- Forensic and intelligence potential of dust particles
- Chemical contamination of aquifer microbiomes
- Cerebrospinal fluid microbiome of spinal cord injuries
- Macroalgae and their associated microbiomes
- Paediatric Microbiome of Cystic Fibrosis
- Viruses or eDNA for detecting rare flora and fauna?
- Is virus transfer by ticks driving lizard speciation?
- Mobilizing motility for microbiome exploration
- How phage alter the wound microbiome
- Impact assessments in Australian arid environments
- Ecology of beneficial butyrate-producing bacteria
- Large algal viruses and wastewater treatment
- Soil microbiome following biodiversity restoration
- Human fecal viromes during chemo-induced neutropenia
Testing the use of environmental DNA to identify black diversity and sites of white shark occurrence – Sample collection
Charlie Huveneers
Environmental DNA has emerged as an approach for monitoring and tracking cryptic biodiversity, and when coupled with existing monitoring techniques can yield a more complete survey of difficult to montitor environments. Huveneers has proposed to couple environmental DNA with the existing survey techniques of baited remote underwater video systems (BRUVS) to increase biodiversity detection. The project will use eDNA to identify white shark (Carcharodon carcharias) nursery areas as well as provide a more comprehensive biodiversity survery of fishes species in this isolate region. The findings will be used to inform existing monitoring programs and provide crucial habitat data for future management of this threatened species.
Exploring the diversity and ecophysiology of pitcher plant microbiomes in Cape York’s iconic bauxite spring ecosystems – DNA extraction
Julian Beaman
The aim of this study is to investigate microbiome diversity patterns in a carnivorous pitcher plant host (Nepenthes mirabilis) to understanding underlying ecological principles structuring the microbiome. Metabarcoding (16s and 18s) will be used to explore the interactions that occur between Nepenthe’s pitcher plants and their associated microbial communities. Host physiological proceses will be measured in parallel with the microbiome to better link the role of the microbiome to the host’s functioning.
INFODUST: The forensic and intelligence potential of dust particles for counter terrorism and national security – Sample collection
Nicole Foster
The ability to determine where a person or object has been is critical for assessing biosecurity risks, and environmental evidence is increasingly recognized as an emerging source of informative information to this end. Foster has proposed to artificially construct differing dust profiles with the aim of determing useful quantities for detection of unique microbial and geochemical signals. These signals will be used to update the current availability of soil reference data to compile a database that can be used for dust profile mapping in forensic case work and strengthen the geospatial and intelligence capabilities of national defence against terrorism.
Influence of chemical contamination on aquifer microbiomes and identification of TCE and PFAS degraders – Sample collection
James Paterson
Terrestrial subsurface environments are the largest microbial reservois on Earth and are increasingly threatened by chemical contaminants including industrial solvents trichloroethene (TCE), and per- and poly-fluoroalkyl substances. This work aims to understand the effect of PFAS on aquifer microbiomes over time using and identify potential bacterial PFAS degraders. Microbiome profiling will be used to identify rapid shifts in the bacterial community to experimental treatments.
Characterisation of the cerebrospinal fluid microbiome from spinal cord injuries – Experimental Design
James Paterson
Trauma or medical conditions can result in spinal cord injuries (SCI) with 80% of patients experiencing chronic pain as a result and this is linked with changes in the gut microbiomes. A recent discovery that the spinal column fluid is not sterile, as was once believed, makes this a potential site for observing a signature for chronic pain SCI. The aims of this study are to characterise and identify shifts in the cerebrospinal fluid (CSF) microbiome of healthy and SCI individuals, and to determine whether the CSF microbiome is the causative agent in chronic spinal cord pain or an early indicator for chronic pain. The project will use shotgun metagenomic approaches.
Whole genome sequencing of the macroalgae Asparagopsis taxiformis and Asparagopsis armarta and their associated microbiomes – DNA extraction
Jessica Carlson-Jones
Of Australia’s greenhouse gas emissions, livestock contributes approximately 10%. The addition of red algae species (Asparagopsis armata and Asparagopsis taxiformis) to livestock diet has been shown to reduce methane emissions by up to 90%. To understand the mechanism for reduced methan emissions this projects proposes to assemble the whole genome of A. armata for the purpose of identifing genes involved in the reduction of methane. The next aim is to characterize the microbiome using WGS sequencing of A. taxiformis, A. armata and the surrounding water column and to identify optimal coastal areas ofSouth Australia for enhance growth potential.
The Paediatric Microbiome of Cystic Fibrosis – Experimental Design
Jessica Carlson-Jones
The vast majority of CF patients suffer chronic lung infections and altered sputum microbiomes and to date most studies have been conducted on adults; therefore it is unknown if CF infections progress in the same manor as adults. This study will generate a longitudinal paediatric CF metagenome database to develop an understanding for the ‘core’ paediatric CF microbiome and how it differs from adults. Further, this work aims to characterize the taxa and gene function dynamics through time in childena and to compare these dynamics to adults CF patitients. WGS sequencing will be used for analysis the the child CF microbiome.
Viruses or eDNA for detecting rare flora and fauna? – Experimental Design
Jim Mitchell
eDNA has been recognized as a priority method towards to finding evidence for the presence of endangered or elusive species of flora and fauna from an environmental samples. However, several limitations exist for its use. With this proposal, Mitchell will compare eDNA and viral DNA from a freshwater environment to compare and contrast their applications for detecting elusive flora and fauna species. The resulting eDNA and viral metagenomes will be combined into a database for future DNA fragment exploration.
Is virus transfer by ticks driving lizard speciation? – Sample Collection
Mike Gardner and Jim Mitchell
Mechanism at the genetic level for parasite and predator drive diversification and ultimately speciations, remain unknown. This project will examine the extent to which that viruses transferred by ticks onto sleepy lizards drive divergent selection of immunge genes, and whether this transfer can influences speciation. Paired metagenomes of lizard blood and tick-gut viromes will be used to form this analysis and to develop a dataset that will inform future microbiome exploration of host-parasite interactions.
Mobilizing motility for microbiome exploration – Experimental Design
Jim Mitchell
Chemotaxis is a behaviour observed in prokaryotes which has provided fundamental insight into infection, symbiosis, signalling pathways, ion pumping and protein assembly. However, studies general only consider a single species, thus understanding for the multispecies and generational effects on chemotaxis is limited. This study will incorporate both metabarcoding and metagenomics to study motility in rich bacterial communities. The project will track naturally migrating microbial communities along a 1-meter tube to track microbial assembly at taxonomic and genetic levels under continuing competition.
How phage alter the wound microbiome – Experimental Design
Peter Speck and Jim Mitchell
Diabetes often comes with a range of emerging complications, one of which are foot ulcers which often lead to amputation of the limb due to infection and antibiotic resistence. A promising approach to circumvent antibiotic resistance in diabetically induced ulcers is with phage mediations approaches. Metagenomics will be used to determine the genetic selection of S. aureus for the presence of various phage within the ulcer microbiome. This study will determine the rate of resistance to the developed phage cocktail and whether there are any notable biases to a specific phage.
Improving impact assessment in Australian arid environments – Sequencing
Maria Angelica D. Rea
Carbon pools in arid and semi-arid biomes are an important driver of the global carbon cycle. Australia’s arid ecosystems serve as an important global carbon sink which is being negatively affected by climate change. Through the support of experimental design by FAME, this project will determine the metagenome‐derived community structures of these arid and semi-arid biomes, and demonstrate the diversity of genes and pathways representative of microbiome adaptations to environmental stressors in soil. The findings of this study will offer further insights into environmental perturbation and how compounding environmental factors affect the arid soil microbiome.
Investigating ecological associations with beneficial butyrate-producing bacteria in soils and air – Sample collection
Joel Brame
Bacterial genes for butyrate production can be found globally in soils, sediments, waterways, oceans, air samples and more. Butyrate-producing bacteria manufacture butyrate (a short-chain fatty acid essential for human health) from the decomposition of organic fibres and amino acids. Exposure to butyrate-producing bacteria found in soils and sediments are known to postiviley contribute to plant-microbe-animal soil ecosystem. This project aims to to identify which locations across this ecological gradient have greater abundance of genes and bacterial taxa related to butyrate-production. The findings will contribute data for ecological research related to short-chain fatty acids, ecosystem restoration and greenspace design and the human gut microbiota assemblage.
Analysis of large algal virus activity in high rate algal ponds – Sample collection
Ian McDonald
The treatment of wastewater using High Rate Algal Ponds (HRAP) results in the breakdown of organic waste through algal and bacterial growth. The system is well understood in the context of bacterial and algal biology, however there is yet to be research into the different populations of virus-like particle (VLP) interacting within the HRAP environment. The project will determine what the mystery VLP populations’ host relationships are, as well as how these VLPs contribute to the dynamics of the HRAP system.
Shifts in soil microbial community structure and function following restoration in a global biodiversity hotspot - Sequencing
Shawn Peddle
In Western Australia’s Kwongan ecoregion (a biodiversity hotspot), there are ongoing ecosystem pressured from land clearing. Though there are efforts to restore the above ground habitats, the soil microbiota and their interation within the soil system and the above ground biota are often neglected in restoration efforts. Therefore this project will assess changes in soil microbiota structure relative to time since restoration; assess changes in soil microbial functionality across multiple stages of ecosystem restoration; and explore relationships between aboveground plant species compositions, soil physicochemical properties, and the soil microbiome. Results are anticipated to provide innovative recommendations on how to improve soil ecological restoration.
Comparison of human fecal viromes during chemo-induced neutropenia – Sample Collection
Sarah Giles
Chemotherapy treatment causes bacterial and viral dysbiosis, leading to numerous adverse side effects, decreased morbidity, and increased mortality outcomes of patients. Faecal transplants are showing promise in restoring gut microbiome biodiversity and improving patient outcomes. Cancer and the gut microbiome appear to be bidirectional; cancer development may alter the microbiome and alterations in the microbiome may affect cancer progression. While bacterial gut biodiversity in human diseases is well studied, the viral populations, which are an important component of healty gut microbiomes, are poorly understood. This project will identify viral populations and determine their shifts post-chemotherapy. Careful experimental design will yield reference quality genomes for novel viruses that are associated with chemotherapy-induced dysbiosis and cancer progression
