Searching SRA Metadata in the Cloud
This is another in our series of posts on searching things in the SRA. As we noted previously, NCBI has moved most of the SRA into the clouds, which makes searching more convenient.
Let’s search through the SRA Metadata attributes (fields)
We can log into the Google Cloud (but you can also use AWS/Azure as you wish), and run a search for a big query.
Note that the Big Query searches are all using standard SQL and here are some videos explaining SQL
SELECT *
FROM `nih-sra-datastore.sra.metadata`
WHERE acc = "DRR071086";
Suppose we have a whole list of IDs to search?
We can extend this approach using WHERE IN like this:
SELECT *
FROM `nih-sra-datastore.sra.metadata`
WHERE acc in ("ERR10082948", "ERR10082949", "ERR10082950", "ERR10082951", "ERR10082952", "ERR10082953", "ERR10082954", "ERR10082955", "ERR10082956", "ERR10082957", "ERR10082959", "ERR10082960", "ERR10082961", "ERR10082963", "ERR10082964", "ERR10082965", "ERR10082966", "ERR10082967", "ERR10082968", "ERR10082970", "ERR10082971", "ERR10082972", "ERR10082973", "ERR10082974", "ERR10082975", "ERR10082976", "ERR10082977", "ERR10082978", "ERR10082979", "ERR10082980", "ERR10082981", "ERR10082982", "ERR10082983", "ERR10082984", "ERR10082985", "ERR10082986", "ERR10082987", "ERR10082989", "ERR10082990", "ERR10082991", "ERR10082992", "ERR10082993", "ERR10082994", "ERR10082995", "ERR10082996", "ERR10082997", "ERR10083000", "ERR10083002", "ERR10083003", "ERR10083004", "ERR10083005", "ERR10083006", "ERR10083008", "ERR10083009", "ERR10083010", "ERR10083011", "ERR10083013", "ERR10083015", "ERR10083016", "ERR10083017", "ERR10083018", "ERR10083020", "ERR10083021", "ERR10083022", "ERR10083023", "ERR10083024", "ERR10083025", "ERR10083026", "ERR10083027", "ERR10083028", "ERR10083029", "ERR10083030", "ERR10083031", "ERR10083033", "ERR10083034", "ERR10083035", "ERR10083036", "ERR10083037", "ERR10083038", "ERR10083039", "ERR10083043", "ERR10083044", "ERR10083046", "ERR10083047", "ERR10083048", "ERR10083049", "ERR10083050", "ERR10083051", "ERR10083054", "ERR10083055", "ERR10083056", "ERR10083057", "ERR10083058", "ERR10083059", "ERR10083060", "ERR10083061", "ERR10083062", "ERR10083063", "ERR10083064", "ERR10083065", "SRR21081047", "SRR21081048", "SRR21081049", "SRR21081050", "SRR21081051", "SRR21081052", "SRR21081053", "SRR21081054", "SRR21081055", "SRR21081056", "SRR21081057", "SRR21081058", "SRR21081059");
We can search the NCBI k-mer based taxonomy profiles too:
SELECT * FROM `nih-sra-datastore.sra_tax_analysis_tool.tax_analysis` WHERE acc = 'SRR21081434' order by ileft, ilevel
And then finally search for a record by the presence of crAssphage (taxonomy ID: 1211417)
SELECT m.acc, m.sample_acc, m.biosample, m.sra_study, m.bioproject
FROM `nih-sra-datastore.sra.metadata` as m, `nih-sra-datastore.sra_tax_analysis_tool.tax_analysis` as tax
WHERE m.acc=tax.acc and tax_id=1211417
ORDER BY m.bioproject, m.sra_study, m.biosample, m.sample_acc
LIMIT 100
(Note without the limit, this will return 140,864 records!)
But I have thousands of Accessions! What do I do?
OK, let’s take it to the next level!
We are going to create a new table in our Big Query database.
Click on your project name and choose CREATE DATA SET:

This opens a side bar where you can give your data set a name! I called my data sra_searches and because I am using the NCBI SRA data, I want to search in us (multiple regions) (although the SRA data is probably all in Iowa).

Now, expand on your data set and click on the three dots and choose Create Table
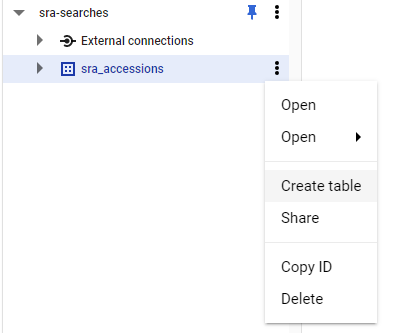
Now we need to fill in four things:
- Choose
Uploadin theCreate Table Frommenu - Choose your file. Use
csveven if you have a single list of accessions! - Give your table a name. e.g.
accessions_20220903(because later I will have other accessions to search!) - Check the
Auto-detecttable format (it works well, I don’t know why it is not the default!)
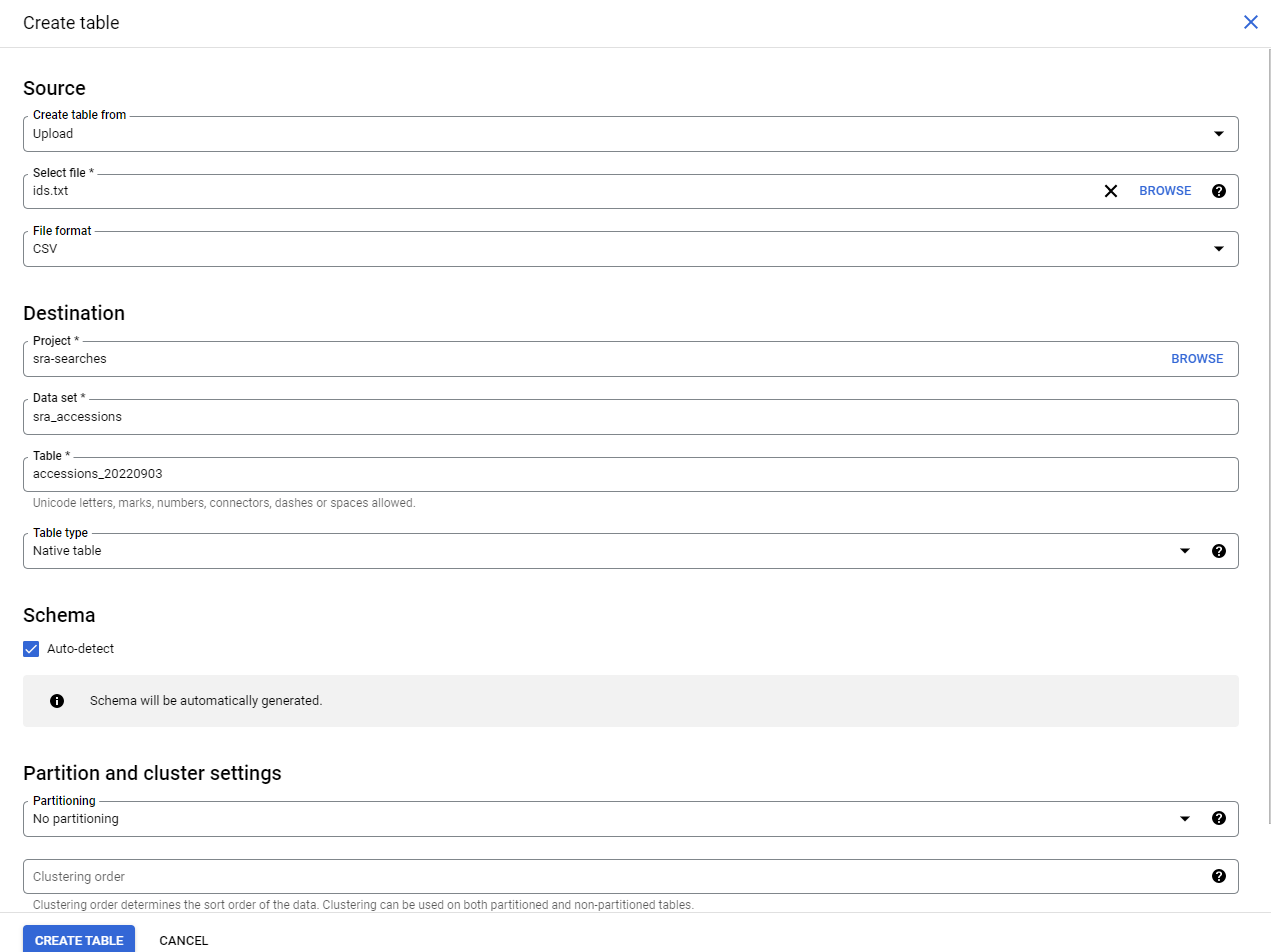
This will create your table, and open it in the browser!
Now click on the data field that has the accessions and the query box will open to allow you to query them:
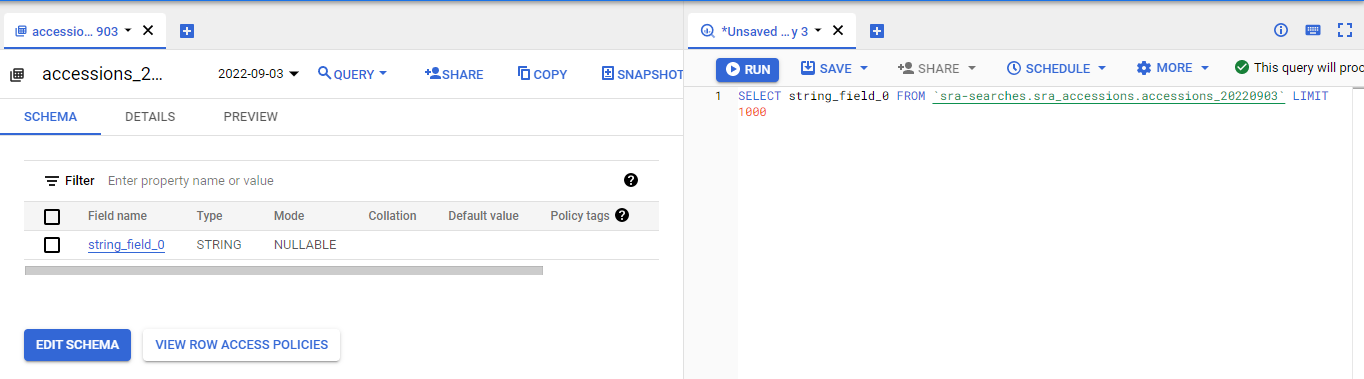
Run that search and make sure it works!
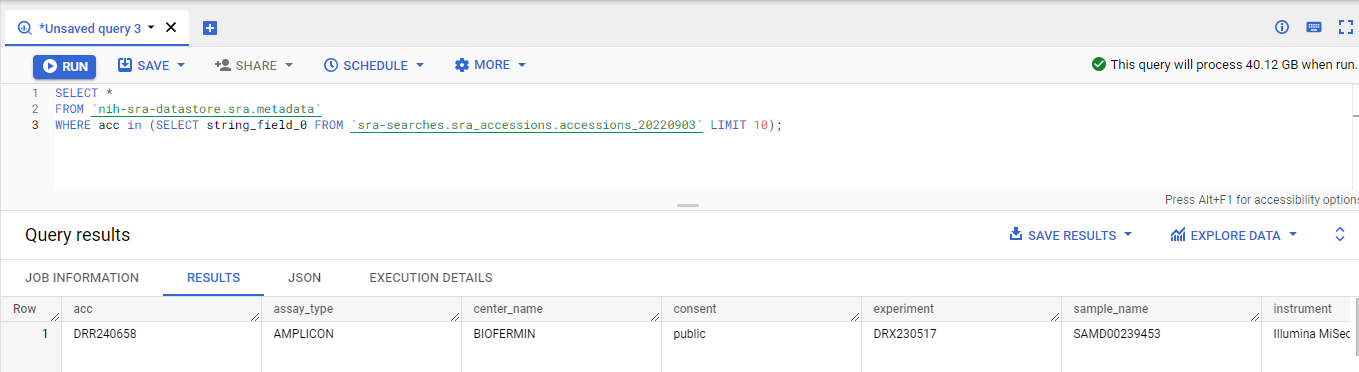
Subselects to the rescue
Now we can just use a sub-select to get all the data.
I recommend
I recommend doing this on a part of your data first to make sure that it works, rather than doing it on all the data!!
Next, I recommend that you export this to Google Drive using JSONL (newline delimited) because it can save upto 1GB:
This makes a format that Google calls JSONL which is not strictly a valid JSON file, because it has one JSON entry per line, whereas a valid JSON file has one entry per file, and would wrap all of this data as a list. However, this format is easy to write on the fly, and much easier to parse if you have a big file as you can read it one line at a time, versus reading the whole file into memory.
We have developed a jsonl2tsv converter that will print the json file as a big tsv file that you can read e.g. into Open Office or Pandas.
